Gut microbiome diversity in a febrile seizure mouse model
Article information
Abstract
Purpose
Febrile seizures at a young age can provoke late-onset temporal lobe epilepsy. Since recent evidence has suggested that the gut microbiome affects central nervous system pathology across the blood-brain barrier, we hypothesized that febrile seizures alter the composition of the gut microbiome to provoke epilepsy.
Methods
Third-generation C57BL/6 mice were separated into two groups (n = 5 each), and hot air was applied to only one group to cause febrile seizures. After two weeks of heat challenge, the fecal pellets acquired from each group were analyzed.
Results
The gut microbiota of fecal pellets from each group revealed five taxa at the genus level and eight taxa at the species level that were significantly different in proportion between the groups.
Conclusion
Although there was no significant difference in the overall diversity of the gut microbiota between the two groups, the identified heterogeneity may imply the pathognomonic causative relevance of febrile seizures and the development of epilepsy.
Introduction
Epilepsy is a common neurologic disease involving more than 70 million people worldwide with a considerable economic burden [1]. The pathophysiological mechanism of epilepsy is characterized by an imbalance between neuronal circuit excitation and inhibition, often attributed to structural defects and inflammatory agents including cytokines and genetic factors [1,2]. Among the various known causes and etiologies, febrile seizures are particularly prevalent in young children, affecting approximately two to four percent of all children [3]. Febrile seizures are considered a significant risk factor for the later development of adult temporal lobe epilepsy (TLE), a correlation supported by a substantial body of epidemiological evidence [4]. It has been determined that, when cortical dysplasia coexists with an immature brain, the likelihood of developing TLE is higher in individuals who have experienced febrile convulsions [5]. However, the exact mechanism underlying the influence of febrile seizures on TLE remains unknown.
Gut microbiota plays a significant role in influencing the physiology and neurochemistry of the central nervous system (CNS), affecting behavior, cognition, mood, anxiety, and depression. Furthermore, the physiology of various neurological disorders such as multiple sclerosis, Alzheimer disease, and Parkinson disease is influenced by gut microbiota [6]. A recent study on the gut microbiome in patients with drug-resistant epilepsy revealed an altered microbiome composition [7]. Another study that analyzed the gut microbiome in status epilepticus-induced TLE rats revealed a distinct metabolic phenotype compared to the control group [8].
We hypothesized that epileptogenesis after prolonged febrile seizure (PFS) could also be influenced by dysbiosis of gut microbiota and analyzed gut microbial composition in a mouse model of PFS.
Methods
The protocols for the animal experiment were reviewed for ethical and scientific care procedures and approved by the Institutional Animal Care and Use Committee of Seoul National University Hospital (No. 17-0158-C1A0). C57BL/6 mice were purchased from Koatech. Second-generation C57BL/6 mice were separated into two groups that each was inbred to produce third-generation mice. Mice born at gestational age 22 to 23 days were designated as postnatal day 1 (P1). At 10 days of age (P10), 10 mice (six males and four females) were injected with 0.2 ml of 0.9% saline to prevent dehydration. Each mouse was placed into a 3-L glass container. Next, all the mice were separated into two groups: a febrile seizure model group (three males and two females) and a control group (three males and two females).
To maintain the core temperature of the test group mice between 41.5 and 42.5 °C, hot air at a temperature of 50 to 52 °C was provided through the upper hole of the cylinder. Mice that became overheated were removed from the cylinder to cool down. The core temperature was measured using a rectal probe every 2.5 minutes during the heating process. Mice that underwent the heating procedures experienced neck jerks, clonic or tonic-clonic seizures, and convulsions. The duration of the seizures was more than 2 minutes and not more than 6 minutes in length. Some of the mice that underwent the heating procedures experienced neck jerks, clonic or tonic-clonic seizures, and convulsions. Mice that experienced seizures during a 45-minute heat exposure were removed from the cylinder during the first seizure, injected with 0.2 mL of 0.9% sodium chloride solution 5 minutes after the seizure started, and allowed to cool for 15 minutes before being returned to their cages. Only mice that had seizures were included in the test group for further evaluation of the gut micriobiome. Seizure events in the mice were observed by two researchers. In contrast, mice in the control group were allowed to move freely within the cylinder to maintain their core temperature at 36 °C. After a 45-minute period of isolation in the cylinder for control purposes, the mice were observed for an additional 15 minutes and then returned to their original cages.
For each mouse, two to three fecal pellets were collected two weeks after the heat shock. These fecal samples were isolated in a deep freezer, their microbiome compositions were determined using metagenomic sequencing of the 16S ribosomal RNA microbial diversity, and differences in the relative abundance of taxa were analyzed by comparing the samples from five mice that underwent PFS induction to those from five age- and sex-matched control mice. For DNA preparation, the FastDNA Spin Kit for Soil (MP Biomedicals) were used. The 16S ribosomal RNA gene V3–V4 region was amplified through polymerase chain reaction using forward and reverse primers (27F: 5’-AGAGTTTGATCMTGGCTCAG-3’, 1492R: 5’-GGTTACCTTTGTTACGACTT-3’). Taxonomic profiling and other analyses were conducted by CJ Bioscience, Inc.
Results
After 2 weeks of daily heat application, analysis of the pellets obtained from each group of mice revealed that the phyla Firmicutes, Bacteroidetes, and Proteobacteria dominated, with no significant differences in abundance between the two groups (Figure 1A). At the class level, in the febrile seizure group, the most prevalent classes were Bacilli, Bacteroidia, and Clostridia in that order (Figure 1B). In the control group, the most prevalent classes were Clostridia, Bacteroidia, and Bacilli, with no significant differences in the abundance of individual classes between the two groups.

Gut microbiome diversity profile of the mice by taxonomic rank
(A) Phylum diversity of the febrile seizure model (case) and control. (B) Class diversity of the febrile seizure model (case) and control. (C) Order diversity of the febrile seizure model (case) and control. (D) Family diversity of the febrile seizure model (case) and control. (E) Genus diversity of the febrile seizure model (case) and control. (F) Species diversity of the febrile seizure model (case) and control.
At the order level, the febrile seizure group exhibited a higher abundance of Lactobacillales, Bacteroidales, and Clostridiales, while the control group had a higher abundance of Clostridia (Figure 1C). Nevertheless, no significant differences were observed in the individual strains at this level. At the family level, there was a decrease in proportion of Lachnospiraceae in the febrile seizure group compared to the control group, but no significant differences were observed (Figure 1D).
At the genus and species levels, significant differences were observed in five and eight taxa, respectively, between the two groups (Figures 1E and F, Table 1). However, when assessing alpha diversity using diversity indices of NPShannon, Shannon, Simpson, and Phylogenetic Diversity, no significant differences were noted between the two groups (Figure 2A). Similarly, alpha diversity assessed using species richness indices of ACE (abundance-based coverage estimator), Chao1, and Jackknife and the number of operational taxonomic units found in microbiome taxonomic profiling exhibited no significant differences between the two groups (Figure 2B). Principal coordinates analysis of beta diversity revealed heterogeneous results between the two groups (Figure 3A). In the linear discriminant analysis effect size analysis, Bacteroides acidifaciens exhibited higher values in the febrile seizure group (Figures 3B and C, Table 1). Taxonomic relative abundance of the known groups, including species Lactobacillus murinus; family Lachnospiraceae; and classes Clostridia, Bacteriodia, and Bacilli, revealed no significant differences between the two groups (Figure 4).
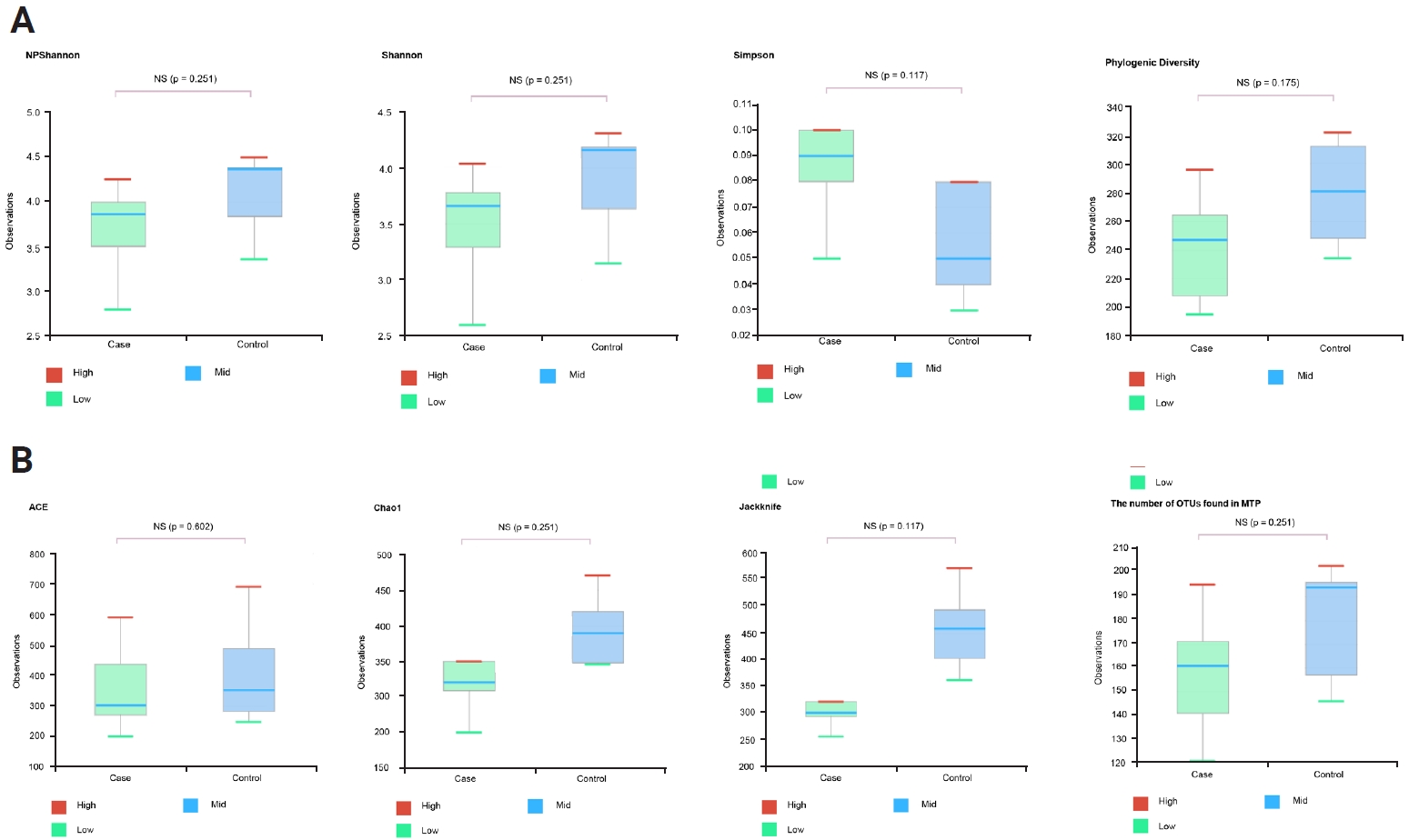
Diversity index (NPShannon, Shannon, Simpson, Phylogenic diversity) and species richness (ACE, Caho1, Jackknife, number of OTUs found in MTP) of the febrile seizure model (case) and controls
(A) There was no significant heterogeneity of diversity index between the groups. (B) There was no significant heterogeneity of species richness between the groups.
ACE, abundance-based coverage estimator; OTU, operational taxonomic unit; MTP, microbiome taxonomic profiling; NS, not significant.

Principal coordinates analysis of the mouse cases and controls
(A) Principal coordinates analysis (Jensen-Shannon, Species, including unclassified OTUs) exhibited significant dissimilarity in beta diversity between the groups (p = 0.049 by PERMANOVA). (B, C) Linear discriminant analysis effect size analysis. Bacteroides caecimuris (B), Bacteroides acidifaciens (C).
OTU, operational taxonomic unit; PERMANOVA, permutational multivariate analysis of variance; PC, principal component.
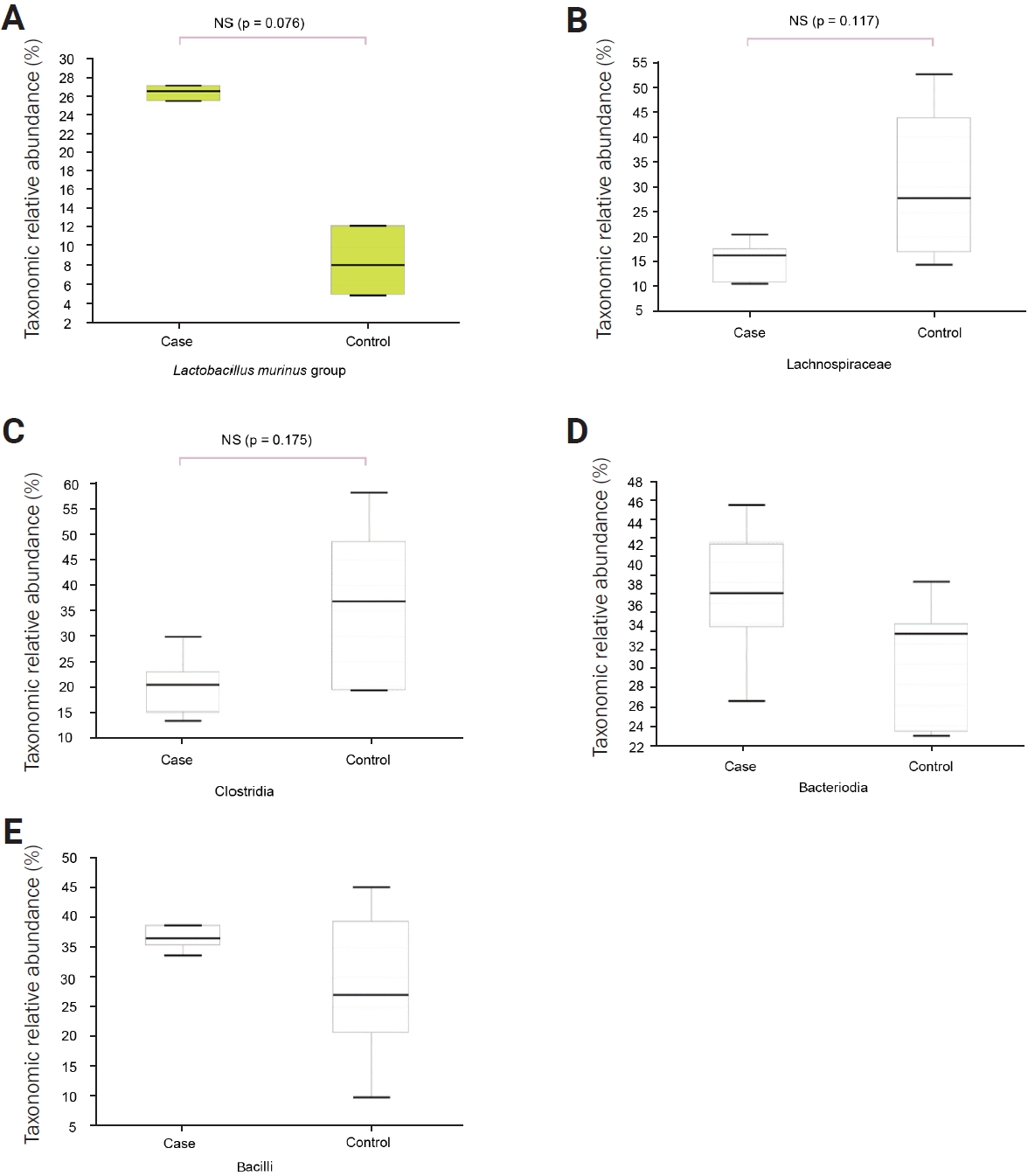
Taxonomic relative abundance of the gut microbiome between the two mice groups
Each panel displays the relative abundance at the representative taxonomic rank. No significant difference in relative abundance was observed between the two groups. (A) Lactobacillus murinus, species. (B) Lachnospiraceae, family. (C) Clostridia, class. (D) Bacteriodia, class. (E) Bacilli, class.
NS, not significant.
Discussion
In our mouse febrile seizure model, taxonomic analysis revealed differences in the composition of gut microbiomes at the genus and species levels compared to the control mice. No differences were observed in alpha diversity, but beta diversity, indicating dissimilarity between the groups, was significant by permutational multivariate analysis of variance (p = 0.049) in eight taxa at the species level and five taxa at the genus level. Bacteroides are recognized for establishing long-term associations with their human hosts and providing health benefits. The two species, Bacteroides caecimuris (p = 0.018) and B. acidifaciens (p = 0.046), exhibited statistically significant differences [9]. The abundance of each identified taxa was quite low, with most less than 0.5%.
Temperature is one of the most important factors for the microbial environment [10]. At a high temperature of 30 °C, the ratio of Bacteroidetes to Firmicutes increased compared to that at 22 °C [11]. High temperature applied to the mouse model in the present study may have altered the composition of the gut microbiome, eventually resulting in an abundance of species including Bacteroides caecimuris and Bacteroides acidifaciens.
Changes in gut microbial composition can also lead to increased gut epithelial permeability, alterations in neurotransmitters within the enteric nervous system, and changes in the CNS. Bacterial microbiota can influence the CNS through pathways such as the vagus nerve and the autonomic nervous system. Vagus nerve stimulation, for instance, can exert anti-inflammatory effects and induce anti-seizure effects. Cytokines, whether proinflammatory or anti-inflammatory, secreted by the gut barrier have the potential to cross the blood-brain barrier and affect both the brain cortex and subcortex, potentially exposing individuals to a higher susceptibility to epilepsy [12,13]. Another review on cerebral cavernous malformation pathogenesis exhibited the role of lipopolysaccharide of gram-negative gut bacteria on the brain epithelium due to a weakened gut barrier [14].
This study has a few limitations. We did not assess the separate role of febrile seizures in the absence of temperature change, which could potentially play a role in triggering epilepsy. An additional study is needed to explore the specific effect of febrile seizure events on the development of epilepsy. Another limitation is the relatively small size of the seizure sample. However, while the sample size may not have been sufficient to draw specific conclusions, the subtle differences in microbial composition observed between the groups underscore the need for further investigation. The gut microbiota has the potential to serve as both a biomarker and a therapeutic target for epilepsy, and further exploration will open new avenues for research.
Notes
Conflicts of Interest
Kyung-Il Park has been an editor-in-chief of encephalitis since October 2020. Daejong Jeon has been an associate editor of encephalitis since October 2020. Keun-Hwa Jung, Soon-Tae Lee, Sang Kun Lee, and Kon Chu have been editorial board of encephalitis since October 2020. They were not involved in the review process of this original article.
Author Contributions
Conceptualization: Jeon D, Park KI, Jung KH, Lee ST, Lee SK, Chu K; Formal analysis, Visualization: Shin YW, Kim Y, Jang Y; Methodology: Shin YW, Jeon D, Yoo J, Park DK, Lee HS, Ahn SJ; Project administration: Shin YW, Yoo J, Park DK; Resources: Jeon D; Supervision: Jeon D, SK Lee, Chu K; Writing–original draft: Kim Y; Writing–review & editing: all authors.

